Package Content¶
The DIAMONDS code is available in a public GitHub repository. The repository, named DIAMONDS, contains the basic package that includes many demos, some of which presented in the main paper of the code, and the modules related to different likelihood functions and prior distributions, useful for a wide range of applications.
Downloading the Basic Package¶
If you want to retrieve the code you have two options:
[Recommended] Open a Git Client (e.g. see SourceTree, available for free). In this case you will have to clone the public repository into your local machine. If you want to clone our public GitHub repository, you need to create a new repository in SourceTree by choosing the option “Clone from URL” and specifying as URL the one of the public repository of DIAMONDS, namely https://github.com/EnricoCorsaro/DIAMONDS. When you clone the repository, we suggest you provide as repository name Diamonds. This option is the best way of retrieving the code because any update, even if small, will always be visible for you using the Git Client. You can then decide to pull the new change, so that the code will be updated without the need to re-download the entire package.
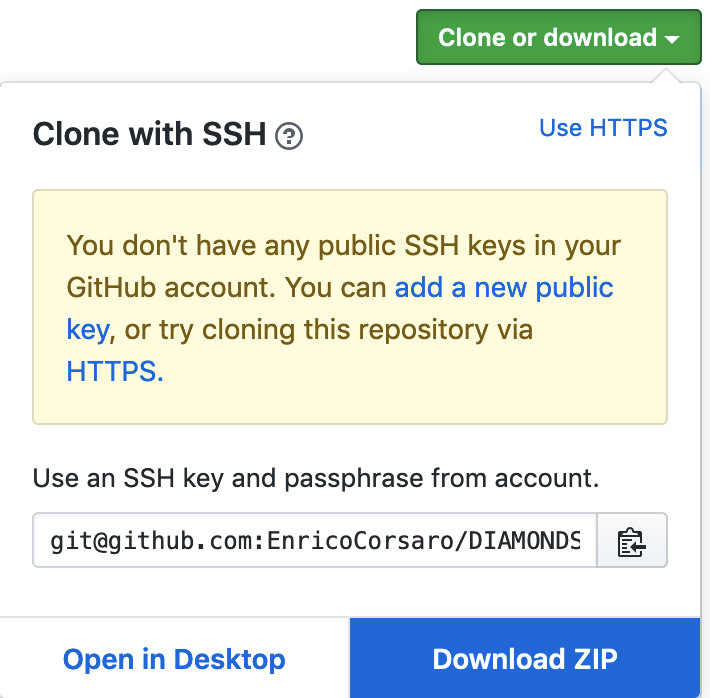
Download the code from the GitHub repository (by clicking on the download as a ZIP file button in the website, see image below). After your download is completed, you unzip the code package file, DIAMONDS-master.zip, and you rename the folder Diamonds-master into Diamonds.
DIAMONDS Basic Package¶
The content of the package is divided into different folders, which are described below:
include: this folder contains the header files of the different C++ classes of Diamonds, namely files having extension .h that contain the class function prototypes.source: this folder contains the source files of the different C++ classes of Diamonds defined in the folder “include”, namely files having extension .cpp that contain the full implementation of all classes functions defined in their corresponding header files.demos: this folder contains a set of demos to test the performance of the code, some of which are presented in the main paper of the code. Each demo has two files with the name coming from the name of the test distribution used in the demo, one with extension .h containing the likelihood function of the demo and its implementation, and the other with extension .cpp containing the main function with the configuration of the Diamonds code and used to run the demo itself. The demos folder also contains a file named mergeMultipleRuns.cpp which can be compiled and used separately for merging multiple runs.tutorials: this folder contains ready-to-use tutorials that can be promptly compiled and executed by the user. These tutorials can be useful as a starting point to implement your own application for parameter estimation and model comparison with DIAMONDS.docs: this folder contains the User Guide manual of the code and the original peer reviewed publication in Astronomy & Astrophysics.
Additionally, four files are also included inside the folder Diamonds. These files are:
CMakeLists.txt. Used for compiling the code files. More information on how to configure the compiling file can be found in the installation guide page.
LICENSE.txt. Contains the license of the code.
README.md. Contains a short description of the code.
improvements.todo. Contains a list of future improvements that we will need to apply to the code.
File Structure¶
We discourage the user, especially if not familiar with the working mechanism of the code, from modifying the content of the source and header files provided in the package. This is to avoid severe impairment of the correct working of the code and of its capability of producing reliable results. For this purpose, the code has been tested to correctly work in the form available by download in this website and any changes to the original files may cause its malfunction.
Header and Implementation files
Header files and their corresponding implementations are divided in different categories, based on the role that the particular class has in the computation process. The categories can be identified by their filename. We use here asterisks legend specifying on which level the user is supposed to change existing classes and/or add new ones:
*** for categories whose classes should never be changed (at the risk to disrupt the correct functioning of the entire code).
** for categories that can in principle be replaced and/or changed but at the risk that computation efficiency may be considerably affected (it is recommended to use those provided in the package).
* for categories whose classes should in principle be usable for the most variety of applications and do not require any change or additions. These categories can however be enriched with new classes for purposes that were not contemplated in the basic version of the code.
[no asterisk] for categories whose classes need to be created and configured by the user based on the problem one wants to investigate.
Sampler***¶
The main class is termed NestedSampler and constitutes the core of the nested sampling computation. The current version of the NestedSampler relies on the usage of a cluster algortihm as a basic algortihm for the sampling. Other implementations that can be provided in the future may include different algorithms than the clustering ones. Derivate classes (concrete) provided in the basic package are:
MultiEllipsoidalSampler- This class implements the Simultaneous Ellipsoidal Sampling algorithm, presented in the main paper of DIAMONDS. This class relies on the use of a clustering algortihm and can in principle be replaced by another sampler class which relies on a cluster algorithm too.ZeroSampler- Empty sampler used in the merger file.
Clusterer**¶
The main class (abstract) is termed Clusterer. Derivate classes (concrete) provided in the basic package are:
KmeansClusterer- The generalized k-means algortihm (X means) for clustering a sample of points, based on the use of the Bayesian Information Criterion.GaussianMixtureClusterer- A Gaussian Mixture model based on Expectation-Maximization algorithm for clustering a sample of points.ZeroClusterer- Empty clusterer used in the merger file.
Reducer**¶
The main class (abstract) is termed LivePointsReducer. Derivate classes (concrete) provided in the basic package are:
PowerlawReducer- A reducer made for removing live points from the nested sampling process to be concentrated toward the end of the computation.FerozReducer- The live points reducer introduced by Feroz et al. 2009 in the first version of MultiNest.
Projector**¶
The main class (abstract) is termed Projector. Derivate classes (concrete) provided in the basic package are:
PrincipalComponentProjector- a Principal Component analysis feature projector to reduce the dimensionality of the inference problem when correlations among free parameters of the model are present.
Metric**¶
The main class (abstract) is termed Metric. Derivate classes (concrete) provided in the basic package are:
EuclideanMetric- Standard Euclidean metric to define the distance between two points in the parameter space.ManhattanMetric- Manhattan metric to define the distance between two points in the parameter space.FractionalDistanceMetric- User controlled fractional distance metric to define the distance between two points in the parameter space.
Likelihood*¶
The main class (abstract) is termed Likelihood. Derivate classes (concrete) provided in the basic package are:
NormalLikelihood- The Normal (or Gaussian normalized) Likelihood used in a wide range of applicationsExponentialLikelihood- The exponential likelihood used in the analysis of stellar power spectra for the study of oscillations (see the original paper of DIAMONDS for an application)MeanNormalLikelihood- A special case of Gaussian Likelihood useful for a more robust treatment of the uncertainties in the observationsMultiLinearNormalLikelihood- A particular form of Normal likelihood to be used for a multi-linear fit, as that shown in the related tutorialZeroLikelihood- Empty likelihood used in the merger file
Prior*¶
The main class (abstract) is termed Prior. Derivate classes (concrete) provided in the basic package are:
UniformPrior- The flat proper prior for a wide range of applications involving the use of simple boundaries on the inferred parametersNormalPrior- The Gaussian normalized prior for cases in which parameter estimates are already knownSuperGaussianPrior- The so-called super Gaussian distribution consisting in a plateau and two Gaussian tails on each side, for mixed situations in which estimates of the free parameters may be known but for preserving a larger degree of confidenceGridUniformPrior- A particular type of distribution which allows to sample only particular ranges of values, each of them contained between regular boundaries equidistant from one another.ZeroPrior- Empty prior used in the merger file
Model¶
The main class (abstract) is termed Model. Among the different categories of classes, this one is the most subject to changes because it is intimately related to the theoretical interpretation one intends to use for the given problem. Derivate classes (concrete) provided in the basic package are
GaussianModel- A model to perform the fit of a multi-dimensional Gaussian as used in the related tutorialSuperGaussianModel- A model to perform the fit of a Super Gaussian modelPolynomialModel- A model to perform the fit of a Polynomial function as used in the related tutorialMultiLinearModel- A model to perform the fit of a multi-linear model as used in the related tutorialZeroModel- used as an empty model in the merger file.
Demos¶
Demo files are provided for testing the correct functioning of the code and can be run immediately after its compilation. Each demo has to be compiled separately from the code and the command line for their compilation is provided at the beginning of each demo .cpp file. When running a demo you assume Diamonds has been compiled already on your machine. The demos were mainly taken from the examples given in the papers by Shaw, J. R., Bridges, M., & Hobson, M. P. 2007, MNRAS, 378, 1365; Feroz, F., Hobson, M. P., & Bridges, M. 2009, MNRAS, 398, 1601; Feroz, F. & Hobson, M. P. 2008, MNRAS, 384, 449; Feroz, F. & Skilling, J. 2013, in American Institute of Physics Conference Series, Vol. 1553, American Institute of Physics Conference Series, ed. U. von Toussaint, 106–113 (FS13).
The demos contained in the basic package are listed below:
2D Single Gaussian
2D Two Gaussians
2D Five Gaussians
Single Gaussian (Any dimensions)
2D Eggbox Function
2D Himmelblau Function
2D Rastrigin Function
2D Rosenbrock Function
2D Two Gaussian Shells
Other demos allow for the testing of the sampling algorithm and of different priors and are:
Prior Drawing 1D - Test the sampling from a given prior in a 1D example
Prior Drawing 2D - Test the sampling from a given prior in a 2D example
Prior Drawing 3D - Test the sampling from a given prior in a 3D example
Kmeans Clusterer 2D - Test the functioning of the Xmeans clusterer from a 2D input sample
Kmeans Clusterer 5D - Test the functioning of the Xmeans clusterer from a 5D input sample
Gaussian Mixture Clusterer 2D - Test the functioning of the GMM-EM clusterer from a 2D input sample
Gaussian Mixture Clusterer 5D - Test the functioning of the GMM-EM clusterer from a 5D input sample
Principal Component Projector 5D - Test the functioning of the PCA projection on a 5D input sample
Ellipsoid 2D - Test the functioning of the ellipsoidal sampler for a 2D example
Ellipsoid 5D - Test the functioning of the ellipsoidal sampler for a 5D example
Ellipsoid 7D - Test the functioning of the ellipsoidal sampler for a 7D example
Input data files for running the demos are all included in the demos folder.
Tutorials¶
The current version of the code includes a number of tutorials that are ready to be compiled and run in your system. These tutorials allow you to have a good starting point to set up DIAMONDS for your own application, as well as providing suitable examples of real-life applications that can be used in a variety of fields. The tutorials available are:
Polynomial Fit: to perform a fit using a polynomial of an arbitrary degree
Gaussian Fit: to perform a fit using a multi-dimensional Gaussian over an arbitrary number of input covariates
Multi-Linear fit: to perform a fit using a multi-linear model over an arbitrary number of input covariates, and related uncertainties
For more information on the tutorials, how to compile the codes, and run the tests, please refer to the hands-on tutorial presentation available here.